My PhD thesis has been published online and can be found at the University of Cambridge repository: https://www.repository.cam.ac.uk/handle/1810/246693
Understanding the epigenome using system genetics
Genetics has been successful in associating DNA sequence variants to both dichotomous and continuous traits in a variety of organisms, from plant and farm animal studies to human disease. With the advent of high-throughput genotyping, there has been an almost routine generation of genome-wide association studies (GWAS) between human disease traits and genomic regions. Despite this success, a particular frustration is that the majority of associated loci are in non-coding regions of the genome and thus interpretation is hard.
To improve characterisation of non-coding regions, molecular assays can be used as a phenotype, and subsequently be used to explain how genetics alter molecular mechanisms. In this thesis, the interplay of three molecular assays that are involved in regulating gene expression is studied. On 60 individuals, several assays are performed: FAIRE-chip, CTCF-seq, RNA-seq and DNA-seq.
In the first part, the discovery and characteristics of FAIRE-QTLs is presented. The identified FAIRE-QTLs show strong overlap with other molecular QTLs, histone modifications, and transcription factors.
The second part consists of the integration of genome-wide molecular assays in a human population to reconstruct the human epigenome. Each of the molecular assays is associated with each of the other assays to discover phenotype-to-phenotype correlations. Furthermore, QTL data are used to dissect the causality for these phenotype-to-phenotype correlations in a system genetic manner.
The third part presents a comprehensive view of CTCF binding on the X chromosome, and its implications for X-chromosome inactivation. A novel X chromosome-wide CTCF effect is observed. Using the gender of each of the cell lines, observations are made about which CTCF sites are dosage-compensated, active on both chromosomes, or are only bound in females.

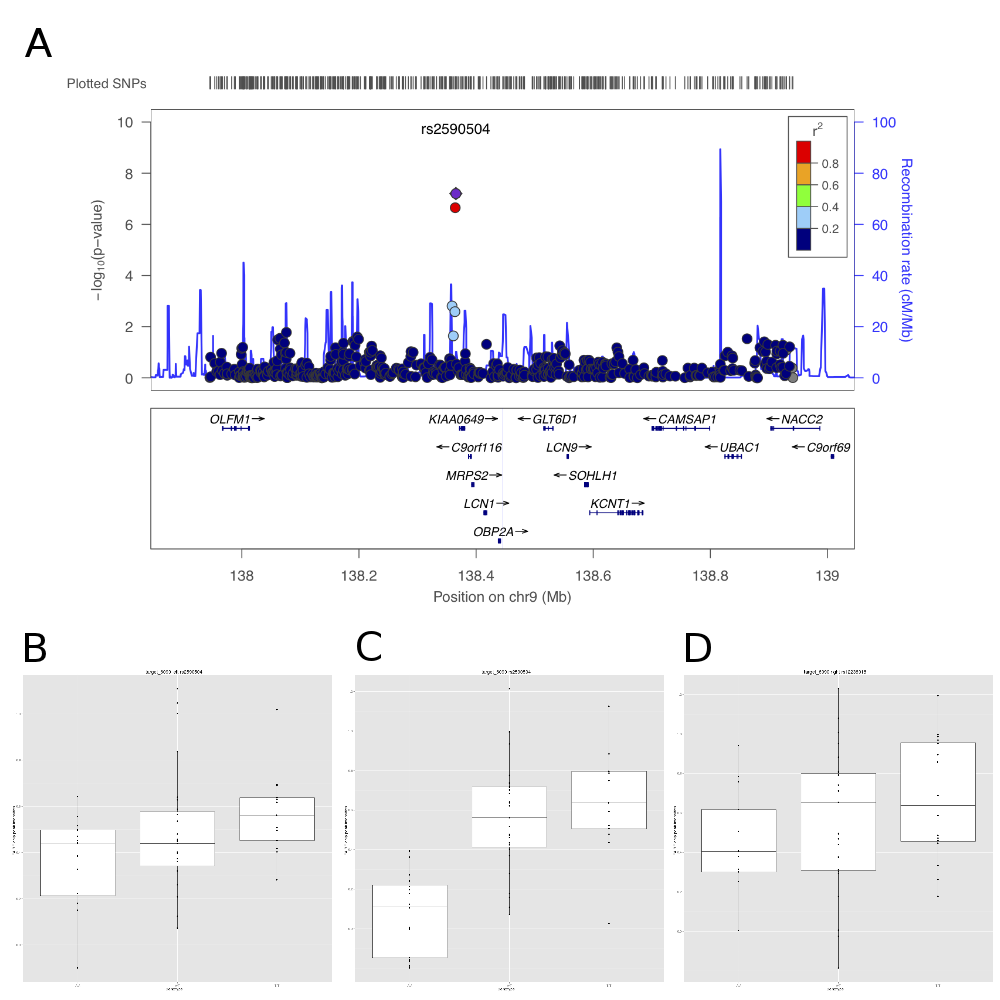
![A. Plot of the metric to distinguish single-active from both active-sites, across the X chromosome for a variety of molecular assays (mRNA, ncRNA, DNase I and CTCF, coloured according to the key). B. A smooth density of the distribution of the dosage compensation fit for the 4 molecular assay types, with CTCF split into the 3 classifications (single active, both active and female specific). C. Allele-specific signal of heterozygote sites on the X chromosome from the 13 clonal female lines in the sample. The both-active sites show balanced allele-specificity, whereas the single-active sites show strong single allele CTCF binding. D. Box plot of the gender-specific behaviour of the DNase I assay at the major classes of X chromosome CTCF sites. DNase I data was collected in a different laboratory on different cell lines [17]. The both-active class shows a strong gender split, consistent with females having around double the signal, whereas the single-active sites show no gender change. doi:10.1371/journal.pgen.1004798.g005](http://www.sandertimmer.nl/wp-content/uploads/2015/01/journal.pgen_.1004798.g005.png)