Reimagining leprosy elimination with AI analysis of a combination of skin lesion images with demographic and clinical data – The Lancet Regional Health – Americas
See the full paper here.
AI4Leprosy: A research project that aims to develop an AI-driven diagnosis assistant for leprosy, based on skin images and clinical data.
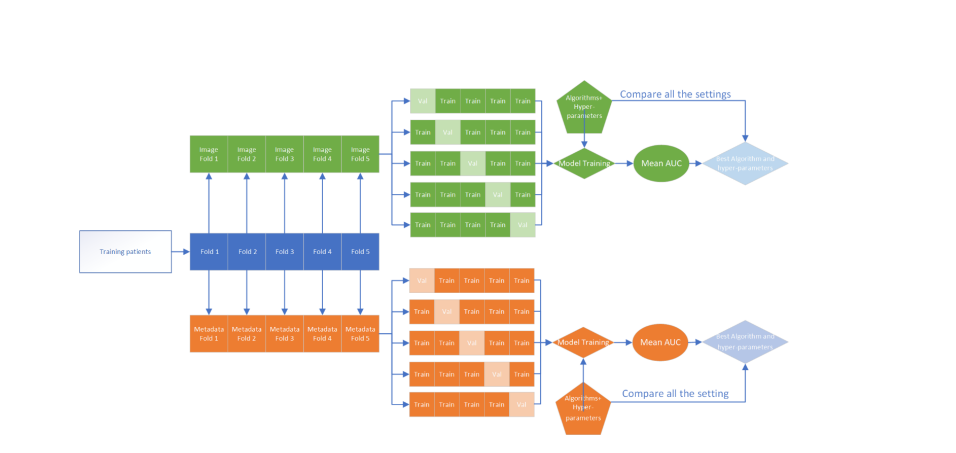
- Dataset: The researchers collected 1229 skin images and 585 sets of metadata from 222 patients with leprosy or other dermatological conditions in a Brazilian leprosy referral center. The dataset is open-source and available for other researchers to use.
- AI models: The researchers tested three AI models, using images and metadata both independently and in combination, to predict the probability of leprosy. They used convolutional neural networks (CNN) for image analysis and elastic-net logistic regression for metadata analysis.
- Results: The best AI model achieved a high accuracy (90%) and area under curve (AUC) of 96.46% for leprosy diagnosis, using a combination of metadata and patient information. The most important clinical signs for leprosy were thermal sensitivity loss, nodules and papules, feet paresthesia, number of lesions and gender.
- Implications: The AI model could be a useful tool to accelerate and improve leprosy diagnosis, especially in low-resource settings. The researchers plan to validate the model in larger and more diverse datasets, and to implement it in a smartphone app for frontline health workers.