Evolutionary indel constraints in intronic regions studied within the 1000 Genomes Project
Minor master summer internship at EMBL – European Bioinformatics Institute, Cambridge, United Kingdom under supervision of Dr. Paul Flicek, Laura Clarke and Dr. Zamin Iqbal.
The 1000 Genomes project is a path-breaking international experiment that involves DNA sequencing on a hitherto unimaginable scale, with the overall goals of generating a baseline understating of the variation in the human genome. In this study we try to detect and understand trends in indel rate in the Human genome. For this project new functionality is added to the Ensembl API to classify the genetic effect caused by indel variation.
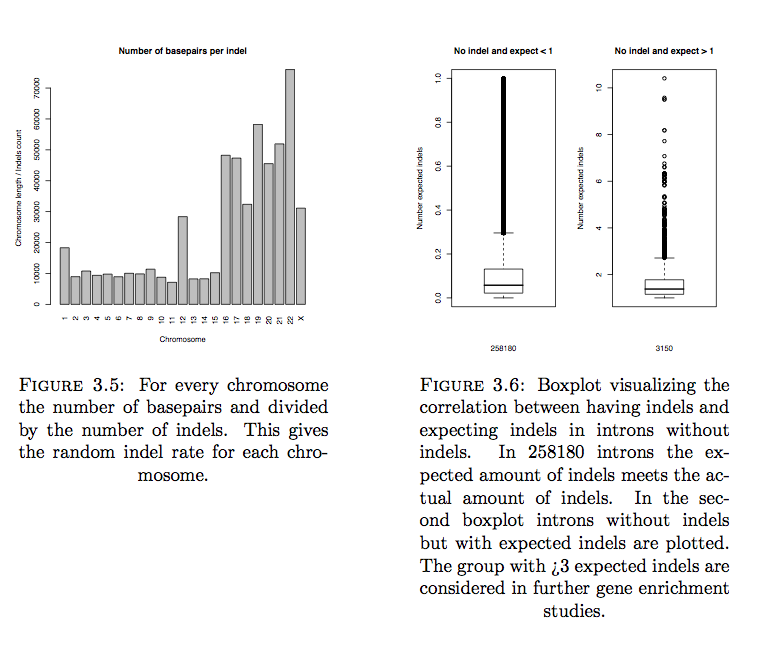
To understand indel rate in Human genome the location and rate of indels are studied. The position of all indels is mapped on the different chromosomes. Genes with and without indels are equally located in the chromosomes (same distance distribution to start/end chromosome). If we look at genes with introns with expected indels we see an overrepresentation in the X chromosome.
Indels were classified in 16 different types of genetic effects. In our trios family sample most indels are located inside intronic regions. Gene enrichment on genes with low indel rate (intron length / number of indels) tend to be involved in RNA binding, which could be evidence for indel constraints in genes with this function.
Next to this I was working for the 1000 Genomes data centrum where I was working on automising sanity checks on the large datasets that had to be mirrored between the different sequencing centers and data centers.